Thermus aquaticus

T. aquaticus is the organism that makes PCR (polymerase chain reaction) possible. It is an ‘thermophile’, capable of living in high temperatures, specifically at temperatures over 70 C (150 F). It was discovered in 1969, at a time when biologists assumed that no living thing could survive at temperatures over 55 C. While Thermus can ‘only’ withstand temperatures up to 80 C, other organisms can live at temperatures even closer to the boiling point of water.
Phylogeny
Unlike many thermophilic (heat-loving) prokaryotes Thermus is not in the Domain Archaea but is a genus in the Domain Bacteria. Along with one other genus Thermus forms a distinct and ancient lineage within the bacteria.
Structure
Thermus aquaticus cells are rod-shaped, non-flagellated gram negative bacteria often occurring as filaments. Gram negative bacteria have a peptidoglycan cell wall layer sandwiched between an inner and outer phospholipid membrane.
Sex and reproduction
Like all bacteria, Thermus are not sexual, but they are capable of exchanging genetic material by other means.
Matter and energy
Thermus is a heterotroph and acquires matter and energy by absorbing organic compounds from its environment, organic compounds that that are derived from other living organisms either by excretion or degradation of the biomolecules that once was part of an organism.
Interactions
Thermus is found in sites with elevated temperatures, hot springs and near thermal vents in the oceans. It occasionally is found in hot water systems and in areas of thermal pollution (e.g. near power plants). It feeds off organic matter produced by other thermophiles including both members of the Domain Bacteria (including some photosynthetic cyanobacteria) as well as members of the Domain Archaea (see Halobacterium)
PCR
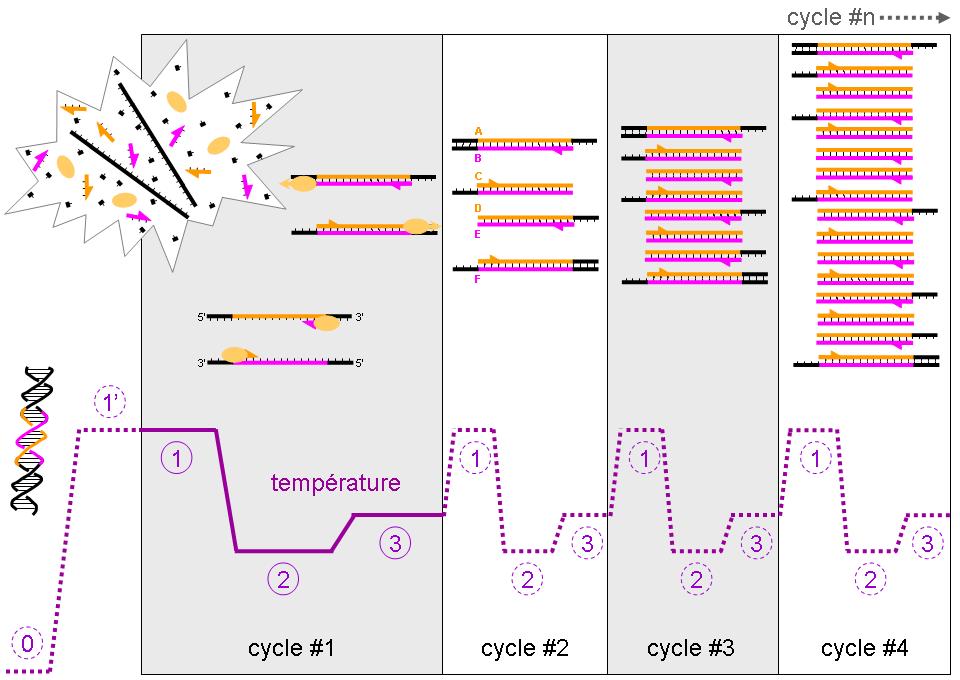
In the last 20 years the PCR technique has revolutionized biology research and plays a very significant role in ‘applied biology’ (e.g. testing for Wuhan flu, paternity testing, diagnosis of hereditary disease, forensic science, security). These are situations where having multiple copies of a certain portion of the DNA molecule are needed and PCR techniques allow the synthesis of multiple copies of a specific part (often a ‘gene’) of DNA. Synthesizing DNA is accomplished by the DNA polymerase enzyme, an enzyme found in all cells. In the normal process carried out by all cells, two enzymes (topoisomerase, helicase) separate a portion of double strand DNA into two single-strands and DNA polymerase is then able to extend DNA strands complementary to each of the single strands that have been revealed. In PCR, heat is used to separate (‘melt’) the double stranded DNA into single strands. Then the mixture is cooled slightly to allow the two ‘primers’ to bind (anneal) to the single strands. The primers are two short sequences of single stranded DNA (one for each strand) complementary to each end of the gene that is to be copied. The annealing of the primers produces a two stranded ‘starting point’ from which DNA polymerase can add nucleotides, thereby extending a DNA molecule complementary to the existing single strand. The DNA polymerase from Thermus aquaticus (called ‘Taq polymerase’) is useful in this process because it can be heated to a temperature high enough to melt DNA yet is still able to function. The PCR machine (called a ‘thermocycler’) performs repeated cycles of high temperature, melting the double stranded DNA, then cooling slightly to allow primer sequences to bind ‘anneal’ to the single strand, and thereby allowing Taq polymerase to work to extend the primer strand in a manner complementary to the single strand. This process (a thermal cycle) is repeated multiple times to get multiple copies of the DNA under study. Although the technique is feasible using polymerases not from thermophilic bacteria, one would have to add additional enzyme after each heating because most enzymes are destroyed by the temperatures required to melt double stranded DNA. Taq polymerase can be added at the beginning and it remains stable through the multiple cycles (usually about 30) needed to produce enough (usually millions!) copies of the gene.
Further Reading
- “Life in Extreme Heat” by Yellowstone National Park Service
- “Yellowstone Hot Springs” by Microbe Wiki
Media Attributions
- Steam from a Hot spring in Yellowstone National Park © Brocken Inaglory is licensed under a CC BY-SA (Attribution ShareAlike) license
- PCR Basic Principle © Ygonaar is licensed under a CC BY-SA (Attribution ShareAlike) license

