Metabolism, Physiology, and Growth Characteristics of Cocci
Metabolism
Like an animal or a plant, the life of bacteria involves a daily routine of thousands of chemical reactions, many devoted to the breakdown (catabolism) of substrates to extract energy or building materials. Other types of reactions utilize the energy and building blocks liberated during catabolism for synthesis reactions (anabolism). The term “metabolism” is an expression used to describe all of the chemical reactions that occur in a cell.
Bacteria rely on enzymes for their biochemistry, just as do other cell types. For bacteria, enzymes needed for metabolic reactions are either endoenzymes, which work within the cell, or exoenzymes, which are produced inside the cell and then transported to the outside where they facilitate the preliminary digestion of high molecular weight substrates that do not pass readily through the cell membrane.
All of this chemistry results in the production of biomolecules and waste, much of which is excreted by the cells into the surrounding environment. Detecting and identifying the biochemical products of metabolism provides us with a way to learn more about the physiologic and growth capabilities of bacteria, and also give us a way to differentiate among and/or identify species.
In Bergey’s Manual, the definitive reference book on bacteria, determinations of identity or taxonomic group are based on many criteria. Gram stain reaction is usually the first criteria, followed by biochemical characteristics such as aerobic or anaerobic respiration, fermentation of various sugars, degradation of proteins and amino acids, and other cellular events. Intermediates or end products of these varied metabolic activities can be detected by performing biochemical assays on a bacterial culture. The results of these tests provide a biochemical profile, or “fingerprint,” that can be used to classify or even identify the bacterial species. The outcomes of laboratory tests can also provide insight into physiology and what is needed to encourage and support bacterial growth.
One of the goals of Koch and Pasteur and their many associates was to develop methods to isolate and identify pathogens as the cause of a human disease. Although the limitations of this approach are now well known, the methods developed during the Golden Age period are still widely used in research and in clinical microbiology laboratories. From clinical specimens, isolated cultures are subjected to a battery of morphological and biochemical fingerprinting tests and compared to known outcomes. In this way, it is possible to identify potentially pathogenic bacteria and distinguish them from the usually helpful symbiotic microbiota.
Morphological Evaluation of Bacterial Isolates
The bacteria we will examine in this lab include species in different genera; Staphylocccus, Micrococcus, Streptococcus, Enterococcus, and Neisseria. At the cellular level, the one characteristic common to all of them is cellular morphology—all are cocci. They differ, however, in many other characteristics.
A volunteer from your lab bench should obtain cultures from your instructor, who will provide you with the species names. Write the name and BSL for each of the cultures below:
_________________________________________________________________________
_________________________________________________________________________
_________________________________________________________________________
_________________________________________________________________________
Colonial morphology
Although there are similarities, the bacteria we will examine in this lab have notable differences, starting with appearance of their colonies. For the bacteria listed in the table below, choose one species from each of the genera below to observe, and describe the colonial morphology of the bacteria in the table below. Note that up until about a decade ago, Streptococcus and Enterococcus were considered part of the same genus and are very similar with regard to both cellular and colonial morphology.
| Bacterium | Colonial Morphology |
| Staphylococcus saprophyticus | |
| Micrococcus luteus | |
| Enterococcus faecalis |
Gram stain and cellular morphology
On a cellular level, all of the bacteria we will look at in this lab have a similar morphology, but there are significant differences in Gram stain reaction, cell size, and cellular arrangements. These differences help to target the particular genus of a bacterial sample.
Prepare smears of the three bacteria you examined (above) and Gram stain them. Also look at a prepared Gram stained slide of Neisseria gonorrhoeae (the causative agent of the STD gonorrhea) and describe what you see in the table below.
Bacterium(write in species name)Gram stain results(reaction, morphology, and arrangement)Staphylococcus _________________(also representative of other Staphylococcus spp.) Micrococcus luteus (also representative of other Micrococcus spp.) Enterococcus faecalis (representative of both Entercoccus and Streptococcus spp.) Neisseria gonorrhoeae(There is no culture of this – view the prepared slide)
Physiology and growth characteristics
Growing bacteria in culture requires consideration of their nutritional and physical needs. Food, provided in the media, is broken down by cells and used for energy and building biomass. Unlike eukaryotic cells, bacteria have options when it comes to making energy, which depend not only on the type of organic molecules in the food but also on the availability of oxygen as a final electron acceptor for respiration.
Respiration is the pathway in which organic molecules are sequentially oxidized to strip off electrons, which are then deposited with a final electron acceptor. Along the way, ATP is made. For many types of bacteria, oxygen serves as the final electron acceptor in respiration. Remarkably, oxygen is not always a requirement for respiration. For bacteria that live in environments with no air, alternative electron acceptors may take the place of oxygen.
Unlike the majority of eukaryotes, bacteria have options when it comes to making ATP. Aerobic respiration and anaerobic respiration generate ATP by chemiosmosis, and some bacteria may also ferment sugars, although the oxidation is not complete and energy is left behind. Chemical by-products and end products of these pathways are detectable and serve as the basis for many biochemical tests performed to identify bacteria.
Fermentation and anaerobic respiration are anaerobic processes—meaning that no oxygen is required for ATP production. Some bacteria have the capability (meaning they produce the appropriate enzymes) to use more than one, or even all three, of these pathways depending on growth conditions.
Based on whether oxygen is required for growth, bacteria can be considered to be either aerobes or anaerobes. However, because some bacteria may use more than one pathway, there are additional categories that describe a culture’s requirement for oxygen in the atmosphere. The three major categories are:
Strict aerobe—Bacteria that are strict aerobes must be grown in an environment with oxygen. Typically, these bacteria rely on aerobic respiration as their sole means of making ATP, but some may also ferment sugars.
Strict anaerobe—These bacteria live only in environments lacking oxygen, using anaerobic respiration or fermentation to survive. For these types of cells, oxygen can be lethal because they lack normal cellular defenses against oxidative stress (enzymes that protect cells from oxygen free radicals).
Facultative anaerobe—The most versatile survivalists there are. These bacteria typically have access to all three ATP-forming pathways, along with the requisite enzymes to protect cells from oxidative stress.
Additionally, overlapping categories include:
Microaerophile—As the name implies, these bacteria prefer environments with oxygen, but at lower levels than normal atmospheric conditions. Often, microaerophiles also have a requirement for increased levels of carbon dioxide in the atmosphere and may also be called capnophiles. These bacteria make ATP by aerobic respiration and may also ferment sugars aerobically.
Aerotolerant anaerobe—These bacteria make ATP by anaerobic respiration and may also be fermentive. However, they are “tolerant” of oxygen because they may have cellular defenses against oxygen free radicals.
Tests that detect either components or end-products of these pathways may be used to assess a culture’s overall oxygen requirement category. The following tests provide the information necessary to assess this growth characteristic.
The catalase test detects the ability of bacteria to produce an enzyme called catalase which is found in cells that live where there is air. Various chemical reactions in electron transport pathways create oxygen free radicals, which are electron-scavenging chemical species that can oxidize and potentially damage biomolecules in cells. One of these is hydrogen peroxide (H2O2), the substrate of the catalase enzyme which converts hydrogen peroxide to water and oxygen. The catalase test is performed by mixing a small amount of a bacterial culture with a drop of hydrogen peroxide on a slide. If the bacteria have the catalase enzyme, the substrate will be split, forming water and oxygen which is observed as bubbling when the gas is released (see Figure 1). A positive test result indicates that the bacteria live aerobically, and are likely to produce ATP by aerobic respiration. Strict aerobes, facultative anaerobes and microaerophiles may be positive for this test. Anaerobes (strict or aerotolerant) will be negative).


The oxidase test identifies bacteria that produce cytochrome oxidase or indophenol oxidases, which are redox enzymes in the electron transport system that shuttle electrons to oxygen. The cytochrome system is usually only present in aerobic organisms that use oxygen as the final electron acceptor in respiration. There are several ways in which this test may be performed, but one of the simplest is to use a commercial test system, such as the BBL DrySlide Oxidase test, which consists of a card saturated with a chemical reagent that is colorless in its reduced state and turns dark blue when oxidized. The cytochrome oxidase enzymes donate electrons to the reagent, changing the color of the card from colorless to blue for a positive test (see Figure 2). Aerobic bacteria with a cytochrome-based electron transport system (similar to what is found in the mitochondria of eukaryotic cells) will be positive for this test.
The nitrate reduction test detects reduced forms of nitrate, which occurs when bacteria use nitrate (NO3) as a substitute for oxygen (O2) during respiration. In the biogeochemical cycle known as the nitrogen cycle, nitrate reduction is the first step in a series of reactions collectively referred to as denitrification (Figure 3).
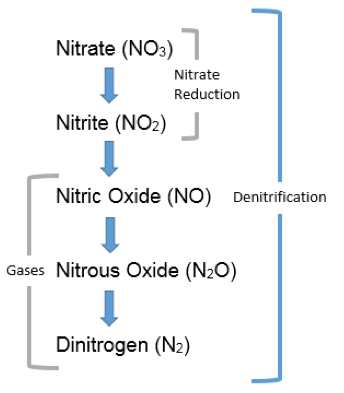
On an ecosystem scale, denitrification decreases the levels of NO3 in soil and slows leaching of this substance into groundwater. On the other hand, denitrification may lead to an increase in N2O, a “greenhouse gas” in the atmosphere and depletes nitrate from soil, which deprives plants and other microbes of this important nutrient. On a cellular scale, some bacteria reduce nitrate as a substitute for oxygen when they are in anoxic environments, and therefore, nitrate respiration can be a useful test for discriminating among bacterial species. This test is performed by subculturing bacteria to nitrate broth, a medium containing food and a source of nitrate available to serve as a final electron acceptor (as a substitute for oxygen for anaerobically respiring bacteria).
Nitrate reduction is demonstrated by adding chemicals that react with nitrite and noting development of a red color, which will occur if the bacteria reduced nitrate to nitrite. No color change after the chemicals are added might mean either the bacteria did not reduce the nitrate at all, or it may also mean the bacteria fully reduced the nitrate to N2 (denitrification). This can be discriminated by adding zinc to cultures that do not change color when the reagents were added. Electrons donated by zinc will subsequently reduce any nitrate remaining in the broth to nitrite, and the broth will become red—therefore a negative test. If the bacteria already reduced all the nitrate to forms other than nitrite, no color change will occur, and this is considered a positive test. A positive nitrate reduction test is indication of an anaerobic lifestyle.
Triple Sugar Iron is a slant medium with two growth environments: aerobic (on the slant) and anaerobic (in the “butt”). The medium contains three sugars in varying concentrations and a pH indicator that turns yellow at pH measurements below 6.8, and a deeper red at pH measurements above 8.2. Bacteria that ferment typically produce one or more types of acid as a byproduct, therefore, fermentation (both aerobic on the slant and anaerobic in the butt) is noted as a change in the color of the media. The medium also identifies strict aerobes that only grow on the slant surface, and also bacteria that produce H2S, either as a way to produce ATP anaerobically using sulfur or sulfate as a final electron acceptor, or as a result of the breakdown of proteins that contain high numbers of sulfur-containing amino acids (cysteine or methionine).
The results of this test are reported as appearance of the slant/appearance of the butt, using A to indicate acid reaction (yellow color), K to indicate an alkaline reaction, and NC to indicate no change in the medium. H2S (detected as a blackening in the media) and the production of gas (CO2) as a byproduct of fermentation are also reported if observed (see Figure 4).
As an example, and for practice, the interpretation and outcomes for the 4 TSI tests shown are provided in the table below. Note that many other possible reactions may also occur so proper interpretation of this test is important.

| Table 1. TSI reactions shown in the cultures in Figure 4, from left to right. | |
| Outcome | Interpretation |
| Uninoculated control | For color comparison with inoculated samples |
| K/NC | Aerobic respiration (dark red on the slant) only. Bacteria are strict aerobes. |
| A/A; gas | Fermentation of all three sugars with CO2 produced. Bacteria are facultative anaerobes. |
| K/A; H2S | Aerobic respiration (dark red on slant), fermentation of glucose (acid only in butt), anaerobic respiration (black in butt). Bacteria are facultative anaerobes. |
| K/A | Aerobic respiration (dark red on the slant); fermentation of glucose (acid only in butt). Bacteria are facultative anaerobes. |
How would you interpret the outcome of the TSI slant, the appearance of which is described below?
| Appearance | Outcome and Interpretation |
| Slant is a dark red color, butt is yellow with noticeable cracking and bubbling. |
After inoculation and test procedures have been demonstrated, perform these tests on the bacteria listed in the table, and record the outcomes below:
| Bacterium | Catalase | Oxidase | Nitrate Reduction | TSI |
| Staphylococcus (aureus OR epidermidis) | ||||
| Micrococcus luteus | ||||
| Enterococcus faecalis |
For each bacterium, determine if the test results provide evidence of aerobic or anaerobic respiration or fermentation, and indicate why you reached that conclusion. Then, based on your observations, state the logical growth category related to oxygen for each.
Staphylococcus ______________(write in species you tested)State what evidence from test results indicates the bacteria use this pathway. If there is no evidence, write “none.”
| Aerobic respiration | |
| Anaerobic respiration | |
| Fermentation |
Oxygen Growth Requirement Category______________________________________________
| Micrococcus luteus | State what evidence from test results indicates the bacteria use this pathway. If there is no evidence, write “none.” |
| Aerobic respiration | |
| Anaerobic respiration | |
| Fermentation |
Oxygen Growth Requirement Category______________________________________________
| Enterococcus faecalis | State what evidence from test results indicates the bacteria use this pathway. If there is no evidence, write “none.” |
| Aerobic respiration | |
| Anaerobic respiration | |
| Fermentation |
Oxygen Growth Requirement Category______________________________________________
Differentiating Among Bacterial Species Based on Phenotypic Characteristics
All of the “volunteer” bacteria used for this experiment are in Bergey’s Group 17 (Gram-positive cocci). From a medical perspective, some are considered primary or opportunistic pathogens, and others are nonpathogens. To distinguish among the cocci in this group, preliminary colonial and cellular characteristics along with growth patterns may be applied, as illustrated in the dichotomous key in Figure 5.
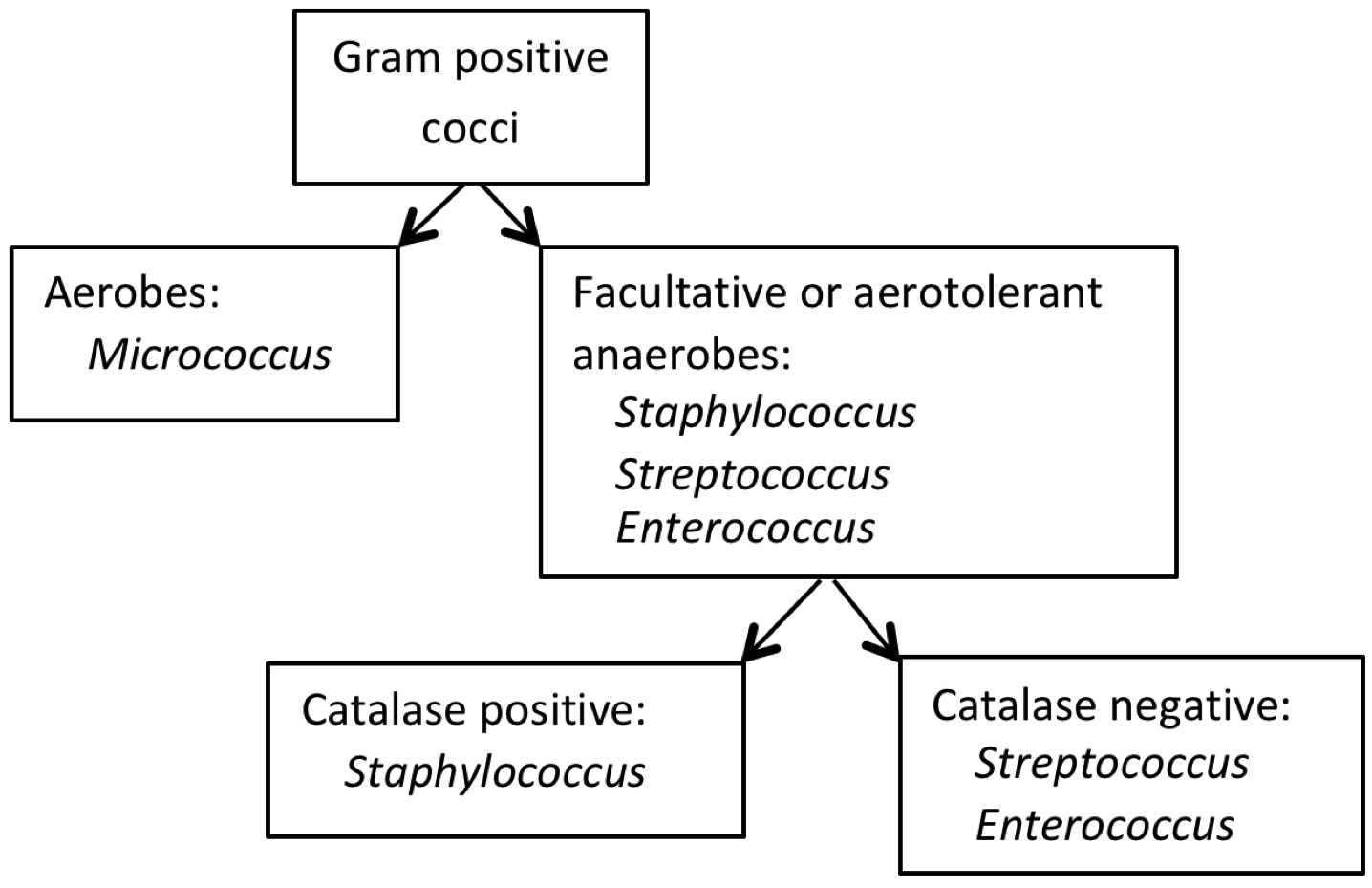
Differentiating among Staphylococcus spp.
There are a large number of laboratory tests that facilitate differentiation among individual Staphylococcus spp. Cultures of three species of Staphylococcus have been provided. The following tests permit the differentiation of these species from one another. Note that an overnight incubation period is required for completion of these tests.
Coagulase test for differentiating S. aureus from other staphylococci
Staphylococcus aureus is known to cause several types of disease in human in addition to foodborne illness. Staphylococcal food poisoning may result from ingesting food contaminated with either the bacteria or a heat-stable enterotoxin produced by the bacteria.
S. aureus differs from most other species of staphylococci on the basis of its ability to produce the enzyme coagulase, which induces blood clot formation, along with other cell surface antigens such as Protein A. Bound coagulase is referred to as “clumping factor.” Coagulase and clumping factor can be identified using lab tests, and in a clinical laboratory this test is done routinely when Gram positive, catalase positive cocci are isolated from clinical specimens.
Because of the clinical significance, commercial kits are available to detect coagulase activity in bacterial cultures. One such kit is Staphaurex, a rapid test for the detection of clumping factor and protein A associated with Staphylococcus aureus. The kit includes a solution of white beads coated with fibrinogen and IgG, and special reaction cards that make the clumping of the beads obvious. When mixed with the reagent, coagulase-positive staphylococci induce the beads to rapidly form large clumps, which are easily seen against the black background of the card. The degree of clumping has to be interpreted by the observer, and this will be demonstrated in lab.

Mannitol Salt Agar: This is a selective medium for staphylococci and other halotolerant bacteria because the high concentration of salt (7.5%) inhibits the growth of bacteria susceptible to the effects of osmotic stress. In addition, the medium contains mannitol, which is a fermentable substrate for some bacteria, and phenol red as an indicator for acid. For those halotolerant bacteria that can grow on this medium, it is also possible to determine whether or not they ferment mannitol, by looking for a color change from red to yellow in the medium. The test is performed by streaking the bacteria over the surface of an MSA plate and incubating. Positive (left side) and negative (right side) results for mannitol fermentation are shown on the MSA plate in Figure 6.
Hemolysis: Some bacteria are known to produce enzymes that break down phospholipids and cause the cell membranes of red blood cells to rupture. Hemolytic bacteria then scavenge the hemoglobin released from the cell, typically to utilize the iron or other “growth factors” from inside the cell. Hemolysis can be observed by streaking bacteria across the surface of a Blood Agar Plate (BAP), which contains intact red blood cells. The BAP plate shown in Figure 7 is an illustration of β-hemolysis (beta hemolysis), seen as a clear area around the bacterial colonies.
Figure 7 shows a BAP plate with colonies that are non-hemolytic. This is referred to as γ-hemolysis (gamma hemolysis). Not shown is a pattern of hemolysis called α-hemolysis (alpha hemolysis), which is not really a true lysis in that the red blood cell membrane is not ruptured, but merely “bruised.” The hemoglobin, which mostly remains in the cell, is reduced to methemoglobin, which is a green color that can be seen surrounding the colonies growing on the BAP.
Urease: Many bacteria have the ability to hydrolyze urea, and some can do it more quickly than others. The enzyme urease is needed, which hydrolyzes urea to ammonia (a basic substance) and CO2. Urease broth contains two buffers—urea, a tiny amount of food, and phenol red. This test is performed by inoculating bacteria into urease broth and incubating. If they produce urease rapidly, the urea in the broth is hydrolyzed and ammonia raises the pH of the broth. This process is detected by the pH indicator which turns deep pink, which is interpreted as a positive test. A positive (left tube) and a negative (right tube) urease test result are shown in Figure 9.

Obtain cultures of three species of Staphylococcus. Then, perform each of the tests above and record the outcomes in the table below, including both what the result looks like (color change, clear area around growth, etc.) and the interpretation of the outcome (positive, hemolysis, etc.). Note that these tests all require that the bacteria be incubated before the test can be completed.
| Test | S. aureus | S. epidermidis | S. saprophyticus |
| Coagulase by Staphaurex | |||
| Mannitol fermentation | |||
| Hemolysis | |||
| Urease |
From the results of the tests you performed on the three Staphylococcus spp., develop a dichotomous key (in the blank space below) that demonstrates how these tests can be applied to distinguish among the three species of Staphylococcus you tested:
Differentiating among streptococcal species
Chain-forming cocci (streptococci) are common members of the mammalian microbiota and sometimes cause disease. Many species inhabit the oral cavity and upper respiratory tract, while others are found in the GI tract. Philosophically, these two diverse habitats prompted taxonomists to split the GI dwelling streptococci into a separate genus, Enterococcus. Those species that inhabit the mouth remain in the genus Streptococcus. Streptococci in general are aerotolerant anaerobes, and can be distinguished from other types of Gram-positive cocci based on their negative response to the catalase test.
Bacteria in these two genera have many common characteristics. Because of their habitat, enterococci are tolerant to higher concentrations of bile. Bile is a yellow-green compound made up of bile acids, cholesterol, phospholipids, and the pigment biliverdin. It is produced in the liver, concentrated and stored in the gallbladder, and released into the duodenum after food is eaten, where it functions as a biological detergent that emulsifies and solubilizes lipids to help in fat digestion. The detergent action of bile also confers a potent antimicrobial activity. Thus, to survive in such an environment, the enterococci not only withstand the antimicrobial effects of bile, but also play a role in secondary bile metabolism in their host.
Bile Esculin Agar is a medium used to isolate and identify enterococci. This medium contains bile salts, which makes it selective for the bile tolerant enterococci, and esculin, which is an organic compound that some of the enterococci are able to chemically break down to glucose and esculetin. The latter substance combines with ferric ions in the medium, and form a complex which turns the both the colonies and the surrounding medium brownish black.
In a clinical laboratory, the medically important streptococci are identified by serological typing into Lancefield groups, which correlate to types of hemolysis observed when the bacteria are grown on Blood Agar Plates. The Group A streptococci, such as the pathogenic Streptococcus pyogenes, are β-hemolytic, while the Group D enterococci are typically non-hemolytic (γ-hemolysis).
Once the procedure has been demonstrated, subculture the two bacteria below to a Blood Agar Plate and Bile Esculin Agar plate and after incubation, observe the differences between them. Record the results below.
| Bile Esculin Agar | Hemolysis on Blood Agar | |
| Streptococcus pyogenes | ||
| Enterococcus faecalis |
On a separate sheet of paper below, construct a dichotomous key to show how the tests you performed may be used to distinguish among the different cocci we experimented with in this lab.
| Table 2. Summary of Test Methods. | |
| Test | Method |
| Catalase | Transfer bacteria to slide and add H2O2; observe for bubbles. |
| Oxidase | Smear bacteria to DrySlide oxidase card; watch for color change that occurs WITHIN 20 SECONDS. |
| Nitrate Reduction | Transfer 2 ml of Nitrate broth to a sterile culture tube, inoculate with bacteria and incubate. After incubation, add Nitrate A and B reagents – red is positive. IF NO CHANGE, add zinc. Red after zinc confirms negative result. |
| TSI | Stab butt and streak slant of TSI slant; incubate. |
| Coagulase | Place a drop of Staphaurex reagent in a circle on the test card. Add bacteria to the drop and mix; then rock the card. |
| Mannitol Salt Agar | Inoculate MSA plate and incubate. |
| Hemolysis on Blood Agar | Inoculate Blood Agar plate and incubate |
| Urease | Transfer 2 ml of Urease broth to a sterile culture tube; inoculate with bacteria and incubate. |
| Bile Esculin | Inoculate BE plate and incubate. |

